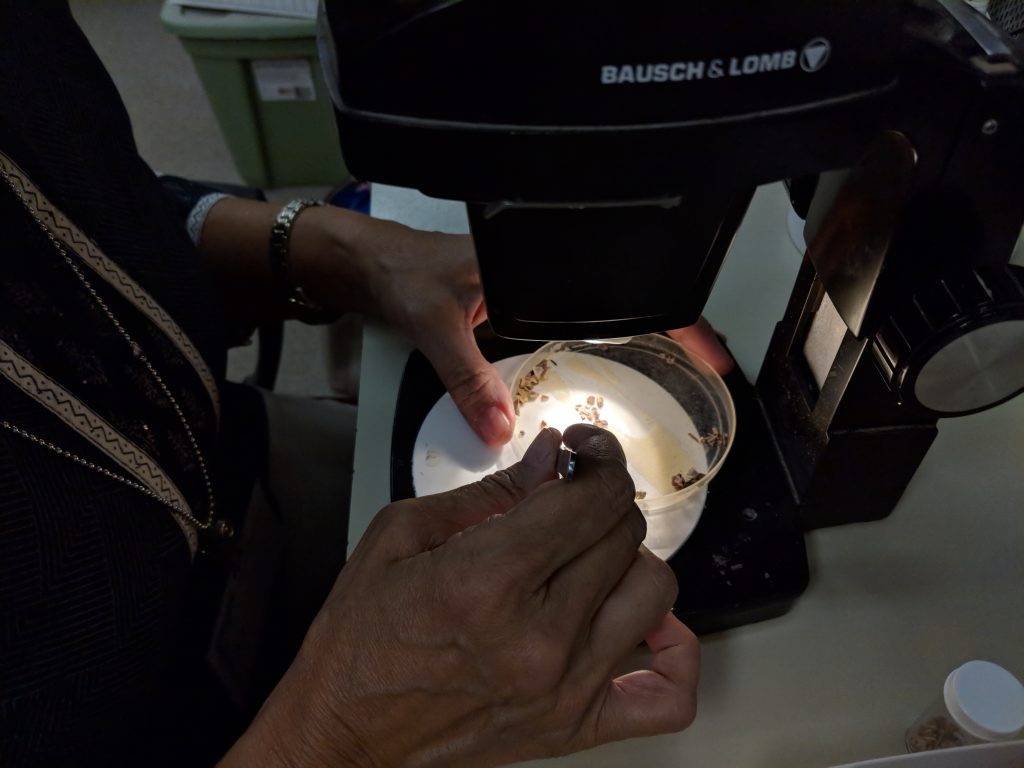
Microvertebrate fossils are really small. They require use of a microscope for identification. Because of this, they receive less love than the huge show off fossils and are often overlooked in the field or discarded with the sediment.

While digging at Montbrook, if we hit a pebbly layer that has fossil colored fragments in it, we’ll bag all of that sediment. We lovingly refer to this layer as, ‘The turtle death layer‘, because it is loaded with turtle fossils.
The bags of sediment eventually make their way back to the museum where volunteers and staff screenwash them in specialized sinks. The bags are poured onto window screens and hosed down so that loose sediment washes out and the fossils and pebbles are left behind. These are placed in cabinets with drying fans and then sent off to the ‘pickers’.
The pickers will sort out the fossils from the pebbles and then further sort the fish from the reptiles and amphibians. We have a much larger sample of fish than reptiles, amphibians, and mammals combined.
These sorted fossils are passed off to the catalogers for further identification and incorporation into the vertebrate paleontology database and collection. This is where it starts to get trickier.
Materials to refer to for identification of microvertebrates, especially fish, reptiles, and amphibians, are sparse.
Plus the fact, that a majority of these fossils are slightly broken. It is common practice to catalog to the nearest order or family without attempting a more precise identification. For instance, with the reptiles and amphibians of Montbrook, we’ve been able to narrow down to:
We may also have some lizards that require further identification.
In order to identify these fossils to a more precise level (most likely to genus), we are utilizing x-ray CT (computed tomography) data made available through the oVert initiative. OVert is working to CT scan every genus of every creature and make these data free to the public! Click the ‘oVert Summary’ button below to learn more.
oVert Summary
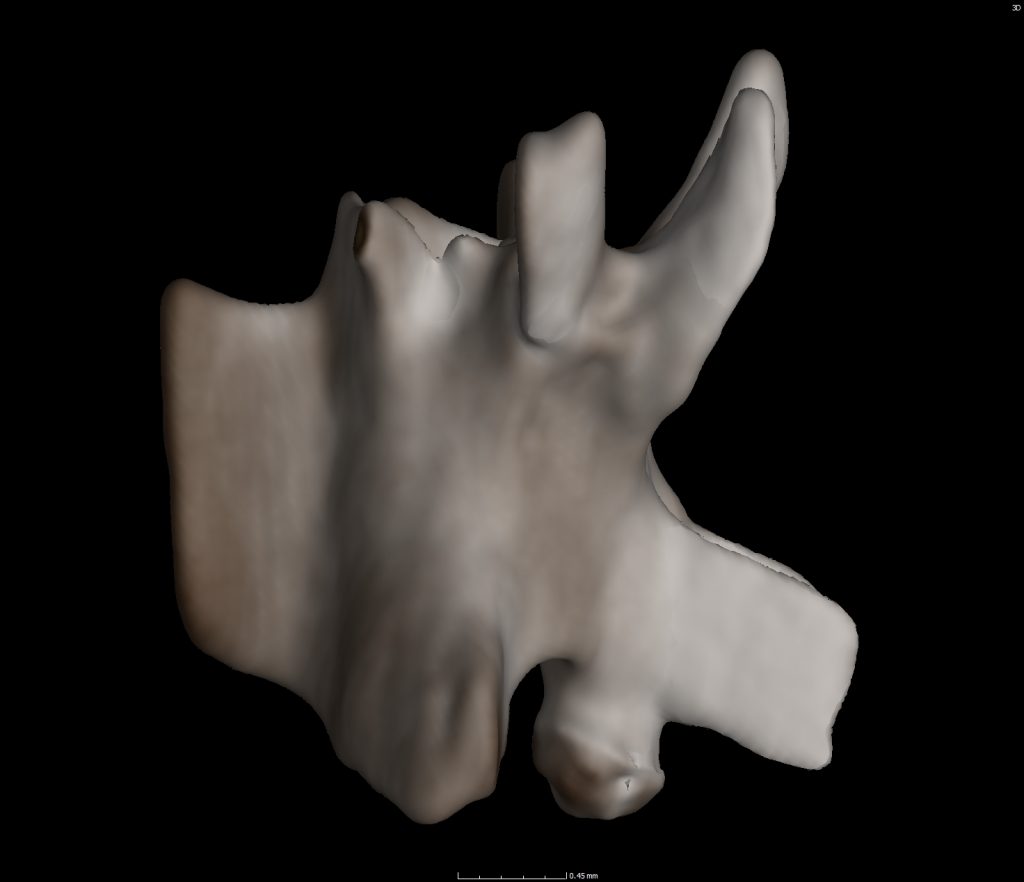
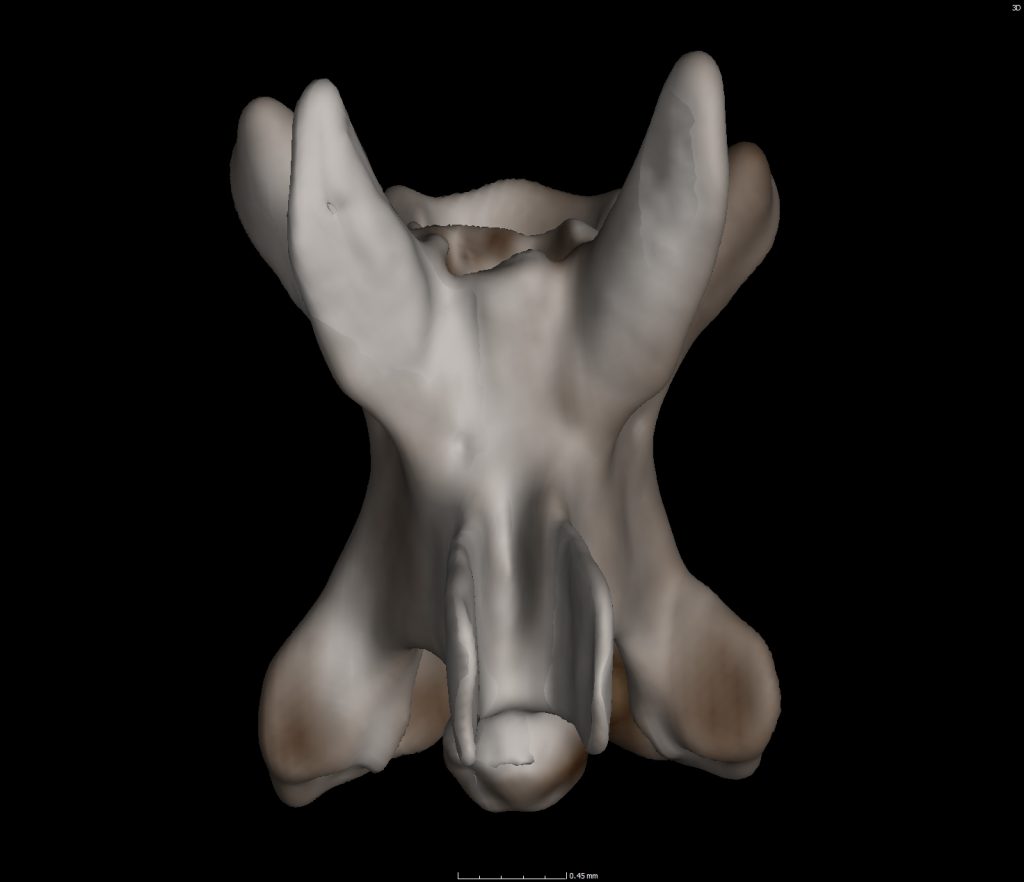
Modern preserved museum specimens, including frogs, salamanders, and snakes are CT scanned to create digitized 3D objects. To create these 3D objects, specimens are placed in the machine on a rotating plate. X-rays from one direction shoot through the specimen and an image lands on the detector plate on the opposite side of the specimen. As the specimen rotates a new image is created. You end up with a stack of images (maybe ~1000 to 2000) that can be combined on specific computer programs (i.e. Volume Graphics, Aviso, etc.) to form a 3D object.
Taking this process a step further, we can utilize those aforementioned computer programs to select a single bone from the modern specimen. We’ve started this process with 4 snakes, 2 salamanders, a glass lizard, and a THUNDERWORM all of which live in Florida; past and present.
A catalog of these bones is being created for our screen washers, sorters, pickers, and catalogers to refer to for greater precision in identification! This also saves time and entices researchers who study specific microvertebrate taxa to conduct science with the Florida Museum vertebrate paleontology collection. A great win for all!
Concluding Thoughts…
These fossils are important because around this time (Late Miocene) many reptiles and amphibians have taken or are taking on their modern appearance. This allows us to directly compare modern reptiles and amphibians to their early forms via examination of the fossils. For some of these species, we’d be able to study the time frame of significant evolutionary changes.
How different are these fossils from the bones of the living relative?
- If they are the same, then this group has not changed much for some 5 million years.
- If they are different, then evolutionary change or events within the family have occurred in the last 5 million years.