In October 2023, I traveled to Friday Harbor, San Juan Island, Washington, to visit the University of Washington’s Friday Harbor Laboratories. After travelling across the country and onto San Juan Island via plane, bus, and boat, I met some of my fellow Computed Tomography (CT) scanning experts from across the United States. We gathered to complete a six-day intensive “train the trainers” 3D Slicer course. 3D Slicer is a free, open-source software for processing CT scan data and is a valuable resource for the scientific community, especially those who don’t have the means to fund expensive CT processing software.

We spent six days under the expert guidance of Murat Maga, Sara Rolfe, Andre Lasso, and Steve Pieper, who first refreshed us on the ins and outs of 3D Slicer, before diving deeply into the software. We learned advanced processing, how to use the python console to perform functions, how new modules are written and implemented into the software, explored our 3D datasets with VR goggles, and how to access a supercomputer (JetStream) capable of dealing with very large datasets.
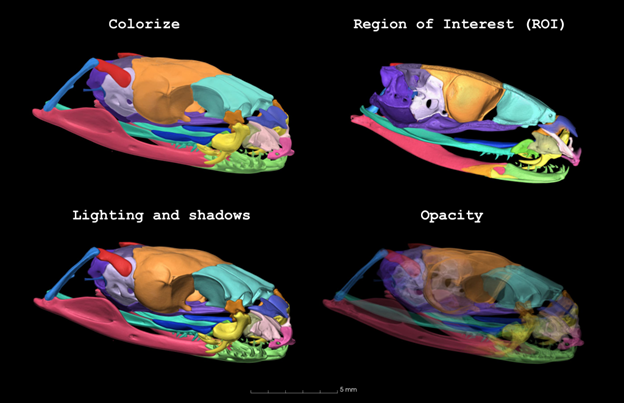 As well as learning from them, we were able to give feedback to the software developers, and after many discussions about the rendering capabilities of 3D Slicer compared to expensive proprietary options, we came up with some ideas for new modules and functions we would like for 3D Slicer. As a direct result of our discussions, a new module called “Colorize Volume” was developed. I am very proud to have played a role in the development of this module, as it has greatly enhanced 3D Slicer’s rendering capabilities.
As well as learning from them, we were able to give feedback to the software developers, and after many discussions about the rendering capabilities of 3D Slicer compared to expensive proprietary options, we came up with some ideas for new modules and functions we would like for 3D Slicer. As a direct result of our discussions, a new module called “Colorize Volume” was developed. I am very proud to have played a role in the development of this module, as it has greatly enhanced 3D Slicer’s rendering capabilities.
At the end of the six days, all of us were inspired to go back to our home institutions and share what we had learned with our colleagues. Now equipped with the knowledge to teach others how to use this software, I have since run several short working group sessions, and will be running a two-day 3D Slicer workshop at the Florida Museum of Natural History in April 2024 with my colleague and 3D Slicer expert Rose Bryson from UF’s Anthropology department.
 After the workshop, I stayed at Friday Harbor for a few more days to get some experience in UW’s Marine Biology lab and visit their CT lab. Hosted by the legendary Adam Summers and my good friend Karly Cohen, I had the most wonderful week. I assisted with two heart surgeries on fish to introduce the contrast agent BriteVu into and 3D image their circulatory system. I worked with Karly Cohen to process Chimeras for her research, and release fish and rays back into the ocean. I was invited on my very first marine biology trawl, where we pulled all kinds of critters out of the ocean to collect and examine. I went “night lighting” where you drop a light into the water at night and observe the weird and wonderful sea creatures who are attracted to it.
After the workshop, I stayed at Friday Harbor for a few more days to get some experience in UW’s Marine Biology lab and visit their CT lab. Hosted by the legendary Adam Summers and my good friend Karly Cohen, I had the most wonderful week. I assisted with two heart surgeries on fish to introduce the contrast agent BriteVu into and 3D image their circulatory system. I worked with Karly Cohen to process Chimeras for her research, and release fish and rays back into the ocean. I was invited on my very first marine biology trawl, where we pulled all kinds of critters out of the ocean to collect and examine. I went “night lighting” where you drop a light into the water at night and observe the weird and wonderful sea creatures who are attracted to it.
And for the cherry on top of a magical trip, it was the perfect time of year to experience the bioluminescence – light emitted by tiny organisms in the ocean.
I am deeply grateful to have received support from the Florida Museum for this trip. Not only was a valuable experience for me, both personally and professionally, but I also returned with skills that will directly benefit members of the FLMNH community who work with CT scan data. The relationships I developed and fostered during the trip will also allow me to have a continuing impact on the development of 3D Slicer and facilitate future improvements to the software that will benefit the wider community.

Jaimi Gray is a postdoctoral researcher at the Florida Museum of Natural History, advised by Dr. Ed Stanley, Associate Research Scientist of the Digital Imaging Division.
The 2023 Fall Student Travel Awards are supported by the FLMNH Department of Natural History, including funds from the Louis C. and Jane Gapenski Endowed Fellowship. If you would like to help support this fund for future student awards, please go to:
Louis C. and Jane Gapenski Endowed Fellowship